Section: New Results
Cells detection using segmentation competition
Participants : Emmanuelle Poulain, Emmanuel Soubies, Sylvain Prigent, Xavier Descombes.
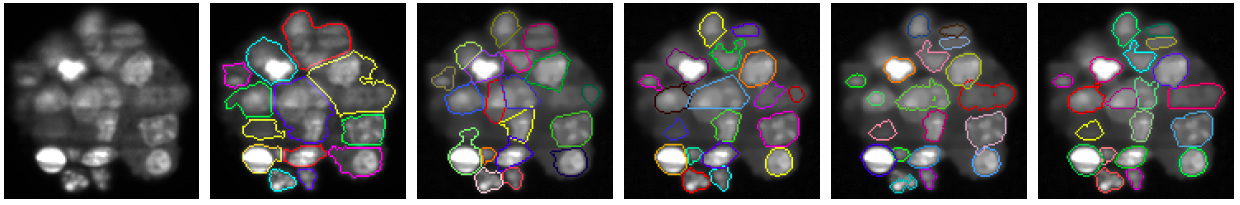
Image segmentation has been widely investigated in particular in the context of bioimaging for cells detection. In some cases, the background is clearly identifiable so that a binary mask of the objects can be computed using simple techniques such as thresholding. Therefore, isolated objects are easily recognizable while splitting clusters of objects, which are connected components in the binary mask, remains a challenging task. In fluorescent microscopy devices used for live imaging – e.g. confocal, biphoton, Selective Plane Illumination Microscope (SPIM) – an additional difficulty comes from the multiple degradations of the acquired images such as strong noise, spatially varying blur and light attenuation which makes the segmentation a hard task even for selecting a suitable threshold for the background. Since many years, researchers have developed several methods to perform such segmentation. An efficient approach consists in generating seeds that define regions using geometric information through a distance, as in the markers controlled watershed algorithm [21] , or image gradient for the active contour approach. These approaches give accurate results providing the seeds are well chosen that is still an open issue. Bayesian approaches, such as marked point process, avoid this bottleneck by selecting randomly generated shapes through the minimization of an energy function. However, they are restricted to low dimensional parametric shapes, such as disks or ellipses, due to computational issues. Tuning the parameters of the segmentation algorithms mentioned above in order to obtain accurate results on the whole image can also be extremely tricky whereas it is much easier to obtain accurate results on different parts of the image using different sets of parameters. To overcome these limits we propose to combine both approaches by generating shapes from state of the art segmentation algorithms using random seeds and/or different sets of parameters. These shapes define a dictionary of candidates from which a competition process, using the Multiple Birth and Cut algorithm, extracts the most relevant shapes. We have validated this selection approach on synthetic data and on a multicellular tumor spheroide slice by comparing the obtained results with two different state of the art segmentation methods to build the dictionary of shapes and compare the performance of our competition approach with the ImageJ particle analyser (see figure 5 ).
|